Deploy and configure CMS claims data transformations (preview) in healthcare data solutions
[This article is prerelease documentation and is subject to change.]
CMS claims data transformations (preview) enable you to use the claims data transformations pipeline to bring your CMS (Centers for Medicare & Medicaid Services) CCLF (Claim and Claim Line Feed) data to OneLake. You can deploy and configure this capability after deploying healthcare data solutions to your Fabric workspace and the healthcare data foundations capability. This article outlines the deployment process and explains how to set up the sample data.
CMS claims data transformations (preview) is an optional capability under healthcare data solutions in Microsoft Fabric. You have the flexibility to decide whether or not to use it, depending on your specific needs or scenarios.
Prerequisites
- Deploy healthcare data solutions in Microsoft Fabric.
- Install the foundational notebooks and pipelines in Deploy healthcare data foundations.
Deploy CMS claims data transformations (preview)
You can deploy the capability and the associated sample data using the setup module explained in Healthcare data solutions: Deploy healthcare data foundations. Alternatively, you can also deploy the sample data later using the steps in Deploy sample data. This capability uses the 8KCCLFClaims sample dataset.
If you didn't use the setup module to deploy the capability and want to use the capability tile instead, follow these steps:
Go to the healthcare data solutions home page on Fabric.
Select the CMS claims data transformations (preview) tile.
On the capability page, select Deploy to workspace.
The deployment can take a few minutes to complete. Don't close the tab or the browser while deployment is in progress. While you wait, you can work in another tab.
After the deployment completes, you can see a notification on the message bar.
Select Manage capability from the message bar to go to the Capability management page.
Here, you can view, configure, and manage the artifacts deployed with the capability.
Artifacts
The capability installs the following two notebooks and data pipeline in your healthcare data solutions environment:
| Artifact | Type | Description |
|---|---|---|
| healthcare#_msft_claims_cclf_extract_bronze_ingestion | Notebook | Converts the raw CCLF data in the Process folder to the respective delta tables in the bronze lakehouse. |
| healthcare#_msft_claims_cclf_fhir_conversion | Notebook | Converts the CCLF data present in the respective delta tables to ExplanationOfBenefit FHIR resources in the bronze lakehouse and saves the output in the form of NDJSON files. |
| healthcare#_msft_claims_cclf_data_ingestion | Data pipeline | Sequentially runs the following notebooks to transform the claims data from its raw state in the bronze lakehouse to a transformed state in the silver lakehouse: • healthcare#_msft_raw_process_movement: Extracts the CCLF files from a compressed (ZIP) file and moves them to the appropriate folder structure based on the execution date in YYYY/MM/DD format.• healthcare#_msft_claims_cclf_extract_bronze_ingestion: Converts the raw CCLF data in the Process folder to the respective delta tables in the bronze lakehouse. • healthcare#_msft_claims_cclf_fhir_conversion: Converts the CCLF data present in the respective delta tables to ExplanationOfBenefit FHIR resources in the bronze lakehouse and saves the output in the form of NDJSON files. • healthcare#_msft_fhir_ndjson_bronze_ingestion: Converts data from the FHIR ExplanationOfBenefit NDJSON file to an ExplanationOfBenefit table in the bronze lakehouse that maintains the raw state of the data source. • healthcare#_msft_bronze_silver_flatten: Transforms data from the ExplanationOfBenefit table in the bronze lakehouse to the ExplanationOfBenefit delta table in the silver lakehouse. |
The healthcare#_msft_raw_process_movement, healthcare#_msft_fhir_ndjson_bronze_ingestion, and healthcare#_msft_bronze_silver_flatten are foundational notebooks that deploy with the healthcare data foundations. To learn more about these notebooks, see Healthcare data foundations: Artifacts.
Set up claims sample data
The sample data provided with healthcare data solutions includes the claims sample datasets to run the claims data transformation pipeline. You can also explore the data transformation and progression through the medallion bronze and silver lakehouses. The provided claims sample data might not be clinically meaningful, but they're technically complete and comprehensive to demonstrate the solution's capabilities.
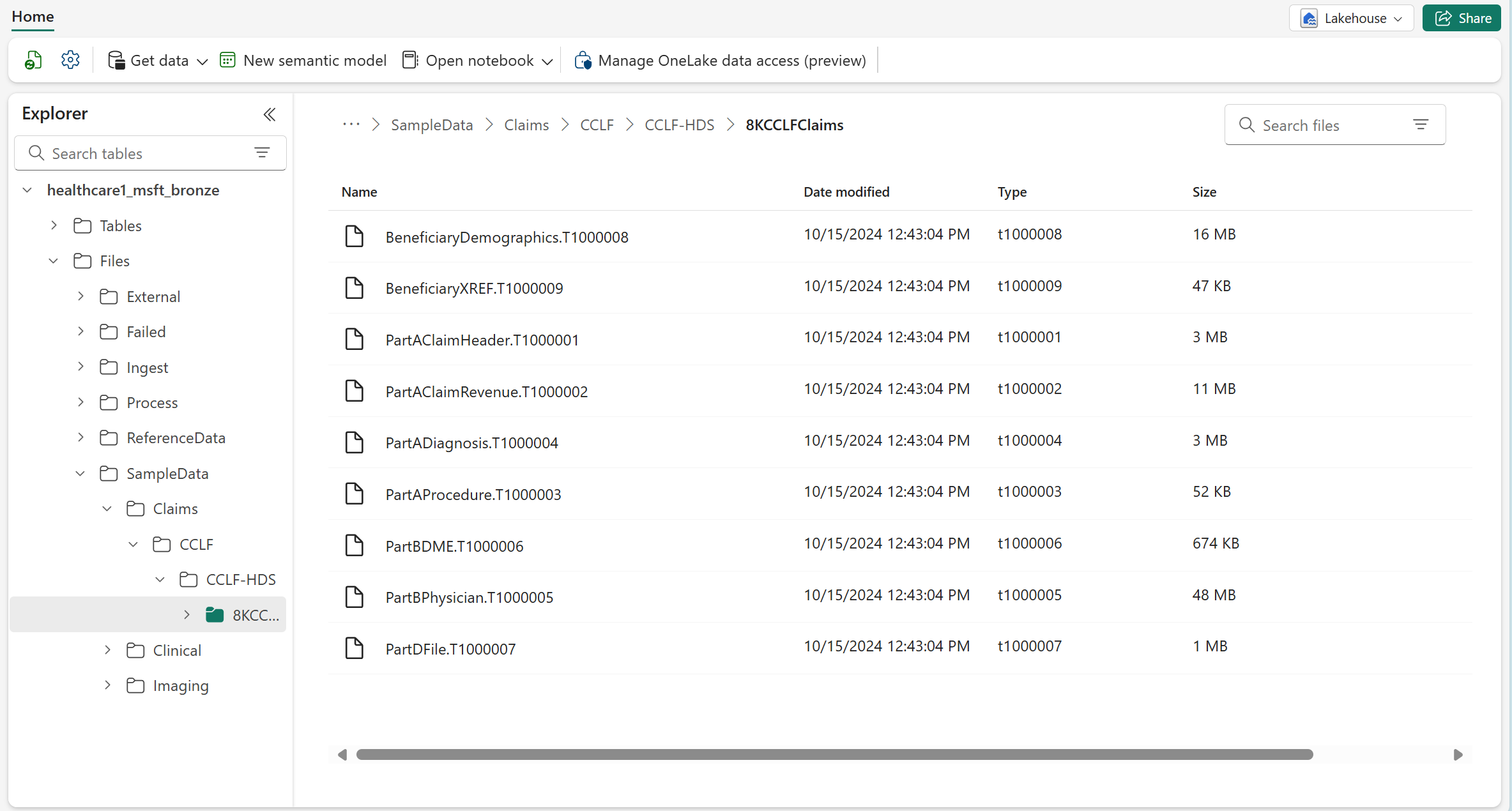
To access the sample datasets, verify whether you downloaded the claims sample data into the following folder in the bronze lakehouse: SampleData\Claims\CCLF\CCLF-HDS. The Deploy sample data step automatically deploys the 8KCCLFClaims sample dataset to this folder.
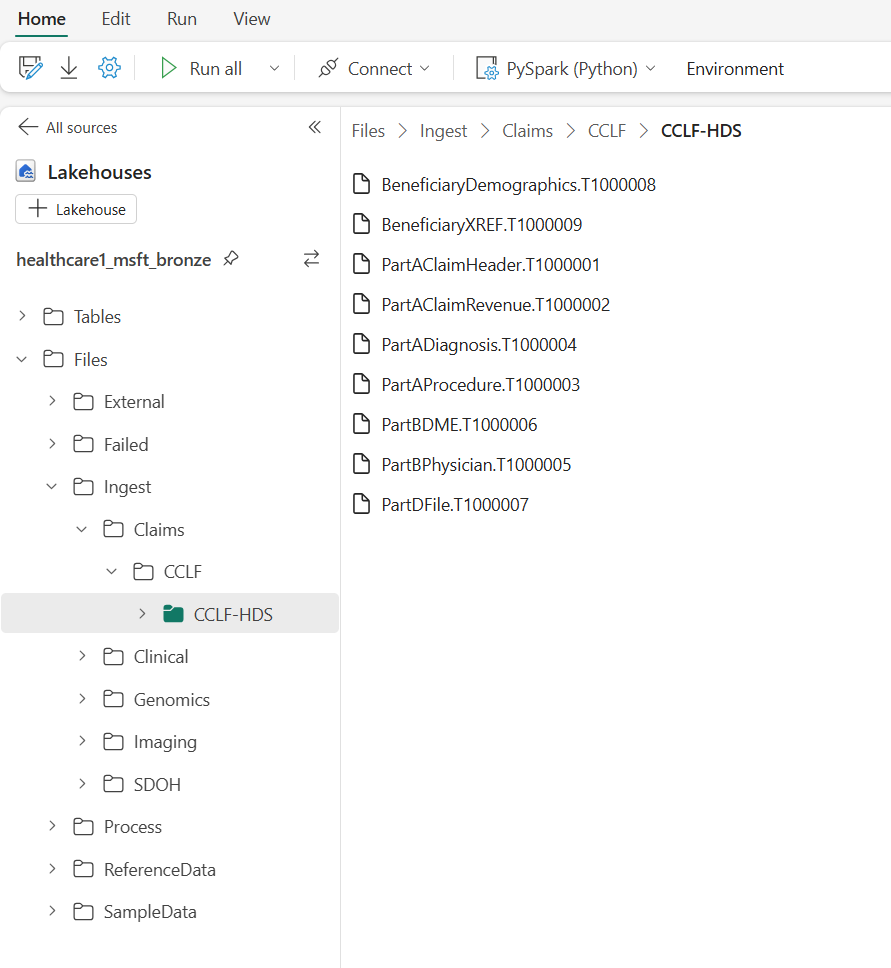
Next, you must upload the sample data to the Ingest folder. This folder in the bronze lakehouse serves as a drop (queue) folder. You must drop the claims sample data files into this folder, so they can automatically move to an organized folder structure within the bronze lakehouse. To learn more about the unified folder structure, see Unified folder structure.
The capability doesn't automatically create the folder path Ingest\Claims\CCLF\<namespace_folder> in your environment. You must manually create this folder path before uploading the sample data.
To upload the sample data:
- Go to
Ingest\Claims\CCLF\<namespace_folder>in the bronze lakehouse. - Select the ellipsis (...) beside the folder name > Upload > Upload files.
- Select and upload the claims dataset from the sample data claims folder.
You can upload either the native claims files (in CCLF format) or ZIP files containing compressed claims files. The ZIP files can include claims files organized into multiple nested subfolders. There's no limit on the number of claims files you can upload or the number, depth, and nesting of subfolders within the ZIP files.
Alternatively, you can run the following code snippet in a notebook to copy the sample data into the Ingest folder.
In your healthcare data solutions Fabric workspace, select + New item.
On the New item pane, search and select Notebook.
Copy the following code snippet into the notebook:
source_path = f"abfss://<workspace_name>@onelake.dfs.fabric.microsoft.com/<bronze_lakehouse_name>/Files/SampleData/Claims/CCLF/CCLF-HDS/8KCCLFClaims" target_path = f"abfss://<workspace_name>@onelake.dfs.fabric.microsoft.com/<bronze_lakehouse_name>/Files/Ingest/Claims/CCLF/CCLF-HDS" files = mssparkutils.fs.ls(source_path) for file_info in files: if file_info.isFile: source_file_path = file_info.path file_name = source_file_path.split("/")[-1] target_file_path = f"{target_path}/{file_name}" try: mssparkutils.fs.cp(source_file_path, target_file_path) print(f"Copied: {source_file_path} to {target_file_path}") except Exception as e: print(f"Error copying {source_file_path}: {e}")Run the notebook. The sample claims datasets now move to the designated location within the folder.