Improving Medical Imaging Diagnostics Using Azure Machine Learning Package for Computer Vision
This post is by Ye Xing, Senior Data Scientist, Tao Wu, Principle Data Scientist Manager, and Patrick Buehler, Senior Data Scientist, at Microsoft.
The advancement of medical imaging, as in many other scientific disciplines, relies heavily on the latest advances in tools and methodologies that make rapid iterations possible. We recently witnessed this first-hand when we developed a deep learning model on the newly released Azure Machine Learning Package for Computer Vision (AML-PCV) and were able to improve upon a state-of-the-art algorithm in screening blinding retinal diseases. Our pipeline, based on AML-PCV, reduced misclassification by over 90% (from 3.9% down to 0.3%) without any parameter tuning. The deep learning model training was completed in 10 minutes over 83,484 images on the Azure Deep Learning Virtual Machine equipped with a single NVIDIA V100 GPU. This pipeline can be constructed quickly, with less than 20 lines of Python code, thanks to the benefit of the high-level Python AML-PCV API.
Our work was inspired by the paper "Identifying Medical Diagnosis and Treatable Diseases by Image-Based Deep learning", published on Cell, a leading medical journal, in February 2018. The paper developed a deep learning AI system to identify two vision-threatening retinal diseases – choroidal neovascularization (CNV) and diabetic macular edema (DME) – and drusen, yellow lipid deposits under the retina, from normal retinal images. The resulting AI system achieved a misclassification rate of 3.9%. Perhaps more importantly, this AI system provides a consistent and objective tool to human experts who may have different opinions on the same image. As a reference, the Cell paper reported that the misclassification rates of the six human experts ranged from 0.3% and 7.9%, with a median at 3.0%. The paper also released the optical coherent tomography (OCT) image sets containing 83,484 images for training and 1000 images from 633 patients for testing. Sample OCT images for these conditions are shown in Figure 1.
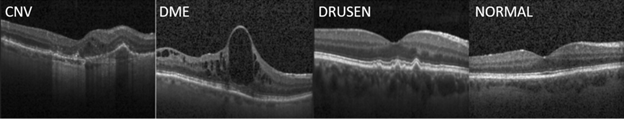
Figure 1: Sample OCT images for retinal diseases
Our objective was to reproduce results from the Cell paper using AML-PCV, and possibly to improve on it. For that, we've used the exact same dataset that authors shared: 83,484 OCT images for training and the 1000 OCT images for testing. In fact, by using AML-PCV, without any parameter tuning, we were able to quickly build the solution and achieve much better results, reducing the misclassification rate from 3.9% to 0.3%.
In our solution, we used a technique called fine-tuning to make a pre-trained model adapt to a new dataset. The idea is first to train a model on a large dataset with millions of images, and in a second step to adjust the weights using the domain-specific dataset. Fine-tuning often benefits domains with relatively small datasets and at the same time carries the accuracy advantage of deep learning. In our case, we used a ResNet50 model pre-trained on millions of natural images from ImageNet as baseline and then used the retinal OCT images to retrain the whole net of the model using AML-PCV.
AML-PCV provides a set of high-level python APIs for computer vision and offers the full power to fine-tune deep learning models. Figure 2 shows the core of our solution, based on AML-PCV. With fewer than 20 lines of code, it includes a complete pipeline of fine-tuning on a pre-trained ResNet50 model. Moreover, with only minor changes to the sample code provided in AML-PCV, we had the solution working on the retinal OCT images. On a machine with NVIDIA K80 GPU, it took approximately one hour to complete the model training. With the new high performance NVIDIA V100 GPU on Azure NCv3 VM, the time for model training is further reduced by over 80%, to about 10 minutes.
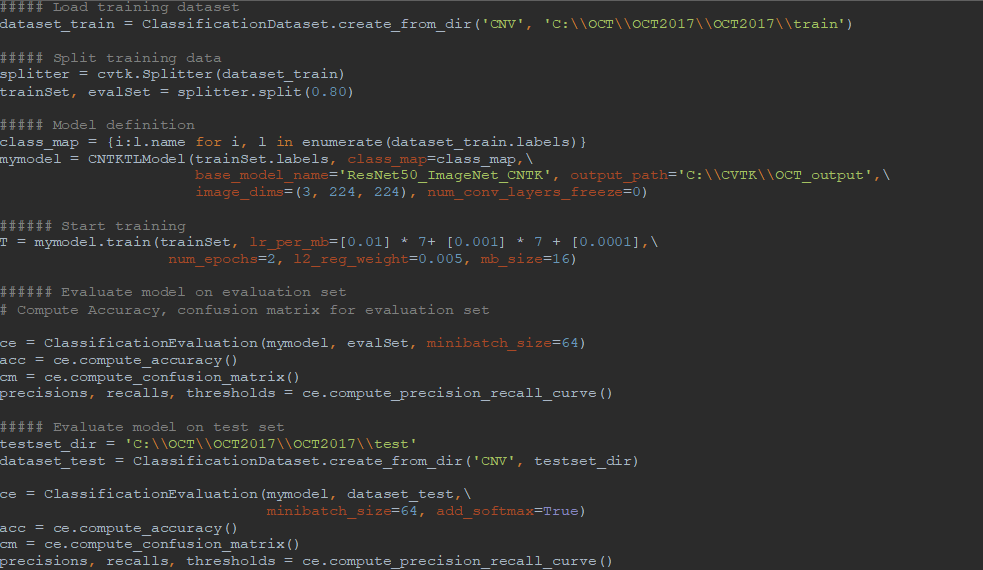
Figure 2: AML-PCV script for retinal OCT image classification
Beyond the great productivity boost, AML-PCV supports state-of-the-art deep learning algorithms and includes default parameters proven to work well on a wide variety of tasks. In our study, our AI system on AML-PCV got a significant performance improvement in classifying retinal diseases compared with that in both AI system and experts reported from the original Cell paper.
More specifically, in the confusion matrix shown in Table 1, among the 1000 OCT images for testing, only 3 images were mis-classified using our model developed by AML-PCV, compared with 39 misclassified images using the AI system from the original Cell paper. It also reaches the same accuracy by the top performer among six experts. Here is a comparison on the confusion matrix of two AI systems between original Cell paper and AML-PCV.
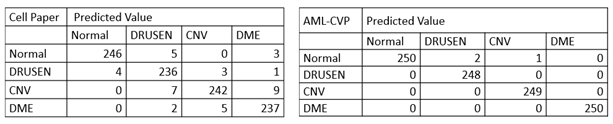
Table 1. Confusion matrix comparison between AI systems reported by original paper and from our study with AML-PCV. The confusion matrix of original Cell paper (left) is based on fine-tuning of last FC layers of Inception V3. The confusion matrix of our study using AML-PCV (right) is based on fine-tuning of whole net of ResNet50.
AML-PCV enabled us to build a working solution in a few hours and achieve better performance than a state-of-the-art AI system. As pointed out in the Cell paper, systems like this can assist doctors to make more robust and objective decisions. On a similar scenario, IRIS, a company started using Microsoft's AI tools to provide primary care doctors with a comprehensive diagnostic platform that allows them to perform the diabetic eye test during a regular check-up, including a medical device, software and services that capture retinal images.
The code for the retinal disease classification task using AML-PCV can be found on GitHub. The code looks simple, but it's nevertheless a very powerful way to achieve state-of-art accuracy in image classification.
Ye, Tao & Patrick