Microsoft Azure ML & R Language Extensibility
How to use External R packages & libraries in MAML
Azure ML studio is Data Scientists’ favorite tool that provides enough functionality for creating and maintaining models for predictive analytics. However, there could be a situation when the existing modules do not suffice for an experiment or, set of requirements. In these cases, ML studio provides the facility of extending the functionality of ML Studio through the R language by using the Execute R Script module. This can also be used in case when you already have your ML module written in R language and you want to import it to ML studio.
This module accepts multiple input datasets and it yields a single dataset as output. You can type an R script into the R Script parameter of the Execute R Script module.
Several R packages are available in addition to the standard packages of the base installation. Currently, it is not possible for you to install R packages directly into the ML studio through the GUI. However, we can install them into the individual workspaces via R code.
A list of the packages included in the current release is provided in the List of installed packages table below.
Listing all currently-installed packages
The list of installed packages can change. To get the complete list, include the following lines in the Execute R Script module send the list to the output dataset:
|
#R code to be used with Execute R Script |
|
|
out <- data.frame(installed.packages()) maml.mapOutputPort("out") |
|
To view the output log, run the experiment, select the Execute R Script module, and click the View output log link near the bottom of the module parameter pane. At this point, we support 400+ R packages out of MAML’s R Engine.
Now, to install & use R language packages which are not supported by the ML studio and are required in your code, we allow the following method.
Here are the key steps:
- Zip up the package(s) to be installed into your workspace for the experiment from R package repository on your machine.
Note: We might need to use multiple packages even in situations where we need only one external library in our code since it might have dependencies which are also not present by default in MAML.
Zip all the zipped packages into another zip file so that all the required packages are bundled together and ready to go
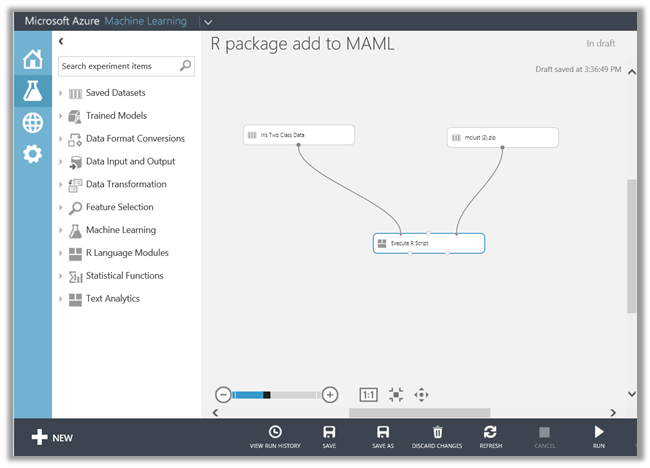
Click on +New at the bottom of the page and upload the zipped file created above as a dataset
Verify that the file has been uploaded successfully
Now, in your experiment add Execute R Script module if it is already not added and connect the dataset input port (if needed) and type/paste your R code which uses the library that we are going to install using the external zipped packages
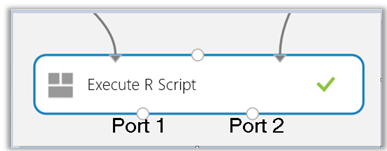
Note: There are 3 input ports on this module
- Drag and drop the uploaded zipped file which contains the packages to be installed
- Use the 3rd input port in the Execute R Script module and connect the zipped file in the previous step
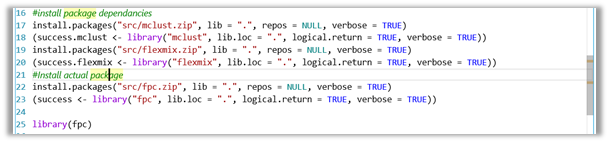
- In the R code just before using the external library which depends on the package(s), use the following code to install the package(s). In this example, I am using the library fpc .
#install package dependancies
install.packages("src/mclust.zip", lib = ".", repos = NULL, verbose = TRUE)
(success.mclust <- library("mclust", lib.loc = ".", logical.return = TRUE, verbose = TRUE))
install.packages("src/flexmix.zip", lib = ".", repos = NULL, verbose = TRUE)
(success.flexmix <- library("flexmix", lib.loc = ".", logical.return = TRUE, verbose = TRUE))
#Install actual package
install.packages("src/fpc.zip", lib = ".", repos = NULL, verbose = TRUE)
(success <- library("fpc", lib.loc = ".", logical.return = TRUE, verbose = TRUE))
#use library
library(fpc,lib.loc = ".")
- Post this run the experiment and use the intended library.
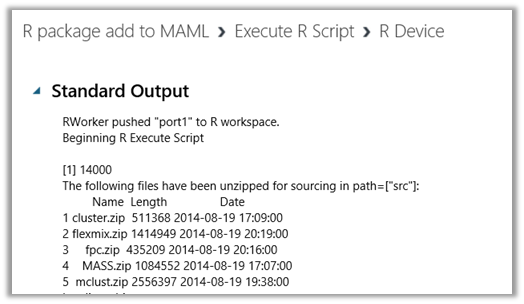
- Once the run is complete, we can look at the output of Execute R Script. Please note that there are two output ports. The first one
- Output port 1 is used to visualize and “Save as Dataset” if there is a dataset output while output port 2 outputs any standard output like verbose output and R plots. In the example here it shows that all the packages which were presented to Execute R Script module from the 3rd input port inside of a zipped file were extracted to be used and placed into the path [“src”].
Additionally, in case you need to use your existing .R and .RData files in MAML, please use the same method as above of zip and upload. Additionally, in the R code inside of the Execute R Script module, please use the following section to provide the names of the files. The zipped file input via the 3rd input port will extract the contents of the zipped file into the path [“SRC”] in the workspace sandbox.
https://channel9.msdn.com/Blogs/Windows-Azure/R-in-Azure-ML-Studio
**Note: Please take into account the legalities of using the R Language Packages while using this functionality.
Comments
- Anonymous
January 01, 2003
Thank you for posting, I was working on an experiment with multiple script bundles this post has helped me. - Anonymous
December 19, 2014
It worked very well for me yesterday today I am getting .zip file not found error, any pointer to what must be going wrong - Anonymous
May 08, 2015
I had to do some trial and error to structure the zip file correctly. Here is what I found:
* Source packages will not work, a Windows binary package has to be used
* If an experiment has multiple R scripts, the packages must be installed in each script that uses one of the packages (it's not possible to start the experiment with a R script dedicated to installing packages, they have to be reinstalled in other scripts)
* Structure of the zip file:
BundleZipFile.zip
|--Package1.zip
|--Package1
|--package files and folders (DESCRIPTION, INDEX, src, R,...)
|--Package2.zip
|--Package2
|--package files and folders (DESCRIPTION, INDEX, src, R,...)
Note that there is no "src" folder in the zip file structure, AzureML will automatically extract to a folder named "src". - Anonymous
May 08, 2015
Correction to previous post (error introduced by comment HTML format):
Structure of the zip file:
BundleZipFile.zip
|----Package1.zip
|--------Package1
|------------package files and folders (DESCRIPTION, INDEX, src, R,...)
|----Package2.zip
|--------Package2
|------------package files and folders (DESCRIPTION, INDEX, src, R,...)