Asynchronous web service consumption via batch processing with mrsdeploy
Important
This content is being retired and may not be updated in the future. The support for Machine Learning Server will end on July 1, 2022. For more information, see What's happening to Machine Learning Server?
Applies to: Machine Learning Server, Microsoft R Server 9.1
In this article, you can learn how to consume a web service asynchronously, which is especially useful with large input data sets and long-running computations. The typical approach to consuming web services, "Request Response" consumption, involves a single API call to execute the code in that web service once. The "Asynchronous Batch" approach involves the execution of code without manual intervention using multiple asynchronous API calls on a specific web service sent as a single request to Machine Learning Server. Then, Machine Learning Server immediately executes those operations once for every row of data provided.
Asynchronous batch workflow
Generally speaking, the process for asynchronous batch consumption of a web service involves the following:
- Call the web service on which the batch execution should be run
- Define the data records for the batch execution task
- Start (or cancel) the batch execution task
- Monitor task and interact with results
Use these following public API functions to define, start, and interact with your batch executions.
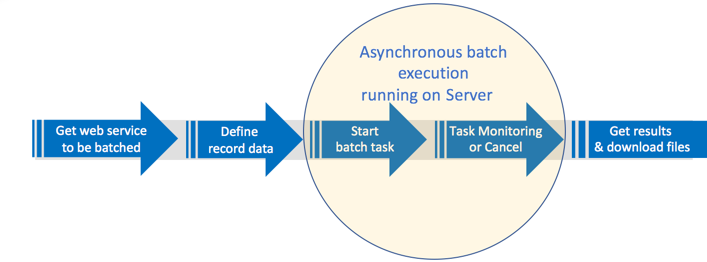
End-to-end workflow example
Use this sample code to follow along with the workflow described in greater detail in the following sections.
Important
Be sure to replace the remoteLogin() function with the correct login details for your configuration. Connecting to Machine Learning Server using the mrsdeploy package is covered in this article.
## EXAMPLE: DEPLOY MODEL & BATCH CONSUME SERVICE ##
##########################################################################
# Create & Test a Logistic Regression Model #
##########################################################################
# Use logistic regression equation of vehicle transmission in the data set
# 'mtcars' to estimate the probability of a vehicle being fitted with a
# manual transmission based on horsepower (hp) and weight (wt)
# Create glm model with `mtcars` dataset
carsModel <- glm(formula = am ~ hp + wt, data = mtcars, family = binomial)
# Produce a prediction function that can use the model
manualTransmission <- function(hp, wt) {
# --- Build a plot to demonstrate files in results ---
png(file = "Histogram.png", bg = "transparent")
hist(mtcars$hp, breaks = 10, col = "red", xlab = "Horsepower",
main="Histogram of Horsepower")
dev.off()
# --- Perdict and return answer ---
newdata <- data.frame(hp = hp, wt = wt)
predict(carsModel, newdata, type = "response")
}
# test function locally by printing results
print(manualTransmission(120, 2.8)) # 0.6418125
##########################################################################
# Log into Server #
##########################################################################
# Use `remoteLogin` to authenticate local admin account.
# Use session = false so that no remote R session started
remoteLogin("http://localhost:12800",
username = "admin",
password = "{{YOUR_PASSWORD}}",
session = FALSE)
##########################################################################
# Publish Model as a Service #
##########################################################################
# Generate a unique serviceName for demos and assign to variable serviceName
serviceName <- paste0("mtService", round(as.numeric(Sys.time()), 0))
# Publish as service using publishService() function from `mrsdeploy`
# package. Use the name variable and provide unique version number.
# Assign service to the variable `api`
api <- publishService(
serviceName,
code = manualTransmission,
model = carsModel,
inputs = list(hp = "numeric", wt = "numeric"),
outputs = list(answer = "numeric"),
artifacts = c("Histogram.png"),
v = "v1.0.0"
)
# Consume service by calling function, `manualTransmission` contained
# in this service
result <- api$manualTransmission(120, 2.8)
# Print response output named `answer`
print(result$output("answer")) # 0.6418125
##########################################################################
##########################################################################
# Perform Batch Consumption & Get Swagger in R #
##########################################################################
# Get the service using getService() function from `mrsdeploy`
# Assign service to the variable `txService`.
txService <- getService(serviceName, "v1.0.0")
# Define the record data for the batch execution task. Record data comes
# from a data.frame called mtcars. Note: mtcars is a data.frame of
# 11 cols with names (mpg, cyl, ..., carb) and 32 rows of numerics.
# Assign to the batch object called 'txBatch'.
records <- head(mtcars[, c(4, 6)], 2) # hp & wt
txBatch <- txService$batch(records)
# Set thread count using parallelCount. Default is 10.
# txBatch <- txService$batch(records, parallelCount = 5)
# Start the batch task
txBatch <- txBatch$start()
# Get the task execution id to reference during or after its execution:
id <- txBatch$id()
# If you need to cancel the batch execution, try this:
# txBatch$cancel()
# Monitor batch execution results with results().
# Check results every 3 seconds until task finishes or fails.
# Assign returned results to batch result object we called 'batchres'.
batchRes <- NULL
while(TRUE) {
batchRes <- txBatch$results(showPartialResult = TRUE) #Default is true
if (batchRes$state == txBatch$STATE$failed) { stop("Batch execution failed") }
if (batchRes$state == txBatch$STATE$complete) { break }
message("Polling for asynchronous batch to complete...")
Sys.sleep(3)
}
# Once the batch task is complete, get the execution records by index from
# the batch results object, 'batchRes'. This object is the service output.
# For every record, return these results (totalItemCount = # of data records)
for(i in seq(batchRes$totalItemCount)) {
#1. List of every artifact that was generated by this execution index.
files <- txBatch$listArtifacts(i)
#2. Get the contents of each artifact returned in the previous list.
for (fileName in files) {
content <- txBatch$artifact(i, fileName)
if (is.null(content)) { stop("Unable to get file") }
}
#3. Download artifacts from execution index to the current working directory
# unless a dest = "<path>" is specified.
# Download of a single named artifact
txBatch$download(i, "Histogram.png")
# Download of all artifacts
txBatch$download(i, dest = getwd())
#4. Get results for a given index row
answer <- batchRes$execution(1)$outputParameters$answer
answer
}
Public functions for batch
You can use the following supported public functions to consume a service asynchronously.
Batch functions performed on the service object
Once you get the service object, use these public functions on that object.
| Function | Usage | Description |
|---|---|---|
batch |
view | Define the data records to be batched and the thread count |
getBatchExecutions |
view | Get the list of batch execution identifiers |
getBatch |
view | Get batch object using its unique execution identifier |
Batch functions performed on the batch object
Once you have the batch object, use these public functions to interact with it.
| Function | Description | Usage |
|---|---|---|
start |
view | Starts the execution of a batch scoring operation |
cancel |
view | Cancel the named batch execution |
id |
view | Get the execution identifier for the named batch process |
STATE |
view | Poll for the state of the batch execution (failed, complete, ...) |
results |
view | Poll for batch execution results, partial or full results as defined |
execution |
view | Get results for a given index row returned as an array |
listArtifacts |
view | List of every artifact files that was generated by this execution index |
artifact |
view | Print the contents of the named artifact file generated by the batch execution |
download |
view | Download one or all artifact files from execution index |
1. Get the web service
Once you have authenticated, retrieve the web service from the server, assign it to a variable, and define the inputs to it as record data in a data frame, CSV, or TSV.
Batching begins by retrieving the web service containing the code against which you score the data records you define next. You can get a service using its name and version with the getService() function from mrsdeploy. The result is a service object, which in our example is called txService.
The getService function is covered in detail in the article "How to interact with and consume web services in R."
Syntax: getService("<serviceName>", "<version>")
Example:
# Get the service using getService() function from `mrsdeploy`
# Assign service to the variable `txService`.
txService <- getService("mtService", "v1.0.0")
2. Define the data records to be batched
Next, use the public api function batch to define the input record data for the batch and set the number of concurrent threads for processing.
Syntax: batch(inputs, parallelCount = 10)
| Argument | Description |
|---|---|
inputs |
Specify the R data.frame name directly, or specify a flat list filename and convert it to a data.frame using the base R function, read.csv. |
parallelCount |
Default value is 10. Specify the number of concurrent threads that can be dedicated to processing records in the batch. Take care not to set a number so high that it negatively impacts performance. |
Returns: The batch object
Example:
# INPUTS = data.frame
# Use mtcars data.frame as input. Assign to batch object 'txBatch'.
# Reduce thread count to 5.
records <- head(mtcars[, c(4, 6)], 2) # hp & wt
txBatch <- txService$batch(records, parallelCount = 5)
# INPUT = Flat CSV converted to a data.frame using read.csv
# Assign data.frame to 'records' variable. Then, use 'records' as input.
records <- read.csv("mtcars.csv")
txBatch <- myService$batch(records, parallelCount = 15)
# INPUT = TSV file converted to a data.frame using read.csv
# Declare the separator
records <- read.csv("mtcars.tsv", sep = "\t")
txBatch <- myService$batch(records)
3. Start, find, or cancel the batch execution
Next, use the public api functions to start the asynchronous batch execution on the batch object, monitor the execution, or even cancel it.
Start batch execution task
Start the batch task with start(). Machine Learning Server starts the batch execution and assigns an ID to the execution and returns the batch object.
Syntax: start()
No arguments.
Returns: The batch object
Example: txBatch <- txBatch$start()
Note
We recommend you always use the id function after you start the execution so that you can find it easily with this id later such as:
txBatch$start()$id()
Get batch ID
Get the batch task's identifier from the service object so you can reference it during or after its execution using id().
Syntax: id()
No arguments.
Returns: The ID for the named batch object.
Example: id <- txBatch$id()
### Get batch by ID
Syntax: getBatch(id)
| Argument | Description |
|---|---|
id |
The batch execution identifiers |
Returns: The Batch object
Example:
service <- getService("name", "version")
# Get a Services existing batch by execution Id
txBatch <- service$getBatch("my-executionId")
print(txBatch)
List the Batch execution identifiers
Syntax: getBatchExecutions()
No arguments
Returns: List of batch execution identifiers
Example: getBatchExecutions()
Cancel execution
Cancel the batch execution using cancel().
Syntax: cancel()
No arguments.
Returns: The batch object
Example: txBatch$cancel()
4. Monitor, retrieve, and interact with results
While the batch task is running, you can monitor and poll the results. Once the batch task has completed, you can get the web service consumption output by index from the batch results object, including:
- Monitor execution results and status
- Get results for a given index row returned as an array
- Get a list of every file that was generated by this execution index
- Print the contents of a specific artifact or all artifacts returned
- Download artifacts from execution index
Monitor execution results and status
There are several public functions you can use to get the results and status of a batch execution.
– Monitor or get the batch execution results
Syntax:
results(showPartialResults = TRUE)
Argument Description showPartialResultsThis argument returns the already processed results of the batch execution even if it has not been fully completed. If showPartialResults = FALSE, then returns only the results if the execution has completed.Returns: A batch result object is returned, which in our example is called
batchRes.
– Get the status of the batch execution.
- Syntax:
STATE
no arguments - Returns: The status of the batch execution.
Example: In this example, we return partial results every three seconds until the batch execution fails or completes. Then, we return results for a given index row returned as an array.
batchRes <- NULL
while(TRUE) {
# Send any results, including partial results, for txBatch task
# Assign it to the batch result variable batchRes:
batchRes <- txBatch$results(showPartialResult = TRUE)
# Check STATUS of the task
if (batchRes$state == txBatch$STATE$failed) { stop("Batch execution failed") }
if (batchRes$state == txBatch$STATE$complete) { break }
# Repeat check every 3 seconds
message("Polling for asynchronous batch to complete...")
Sys.sleep(3)
}
Get execution results as an array
Get the execution results for a given index row.
Syntax: execution(index)
| Argument | Description |
|---|---|
index |
Index value for a given batch data record |
Returns: The execution results for a given index row as an array.
Example: In this example, we return partial results every three seconds until the batch execution fails or completes. Then, we return results for a given index row returned as an array.
# In a loop, get results for a given index row returned as an array in a loop
# assign it to 'exe' and output a data frame for each row.
for(i in seq(batchRes$totalItemCount)) {
answer <- batchRes$execution(1)$outputParameters$answer
answer
}
Get list of generated artifacts
Retrieve a list of every artifact that was generated during the batch execution for a given data record, or index, with listArtifacts(). This function can be made part of a loop to get the list of the artifacts for every data record (see workflow example for a loop).
Syntax: listArtifacts(index)
| Argument | Description |
|---|---|
index |
Index value for a given batch data record |
Returns: A list of every artifact that was generated during the batch execution for a given data record
Example: files <- txBatch$listArtifacts(i)
Display artifact contents
Display the contents of a named artifact returned in the preceding list with artifact(). Machine Learning Server returns the ID for the named batch object.
Syntax: artifact(index, fileName)
| Argument | Description |
|---|---|
index |
Index value for a given batch data record |
fileName |
Name of file artifact created during batch execution |
Returns: The ID for the named batch object
Example:
for(i in seq(batchRes$totalItemCount)) {
for (fileName in files) {
content <- txBatch$artifact(i, fileName)
if (is.null(content)) { stop("Unable to get artifact") }
}
}
Download generated artifacts
Download any artifacts from a specific execution index using download(). By default, artifacts are downloaded to the current working directory getwd() unless a different dest = "<path>" is specified. You can choose to download a specific artifact or all artifacts.
Syntax: download(index, fileName, dest = "<path>")
| Argument | Description |
|---|---|
index |
Index value for a given batch data record |
fileName |
Name of specific artifact generated during batch execution. If omitted, all artifacts are downloaded for that index. |
dest |
The download directory on your local machine. The default is the current R working directory. The directory must already exist on the local machine. |
Returns: The path to each downloaded artifact.
Example:
In this example, we download a named artifact for the fifth index to a specified directory, and then download all artifacts to the default working directory.
#Download a named file for 5th index to the specified directory
txBatch$download(5, "Histogram.png", dest = "C:/bgates/batchfiles/")
# Download all files for a given index to the default current R working directory in a loop
for(i in seq(batchRes$totalItemCount)) {
txBatch$download(i)
}