Sequence Alignment using Concurrency Runtime
Introduction to Sequence Alignment problem domain
Sequence Alignment techniques are very useful in the field of Bioinformatics used for analyzing DNA and Proteins. Sequences are compared and/or aligned to observe patterns of conservation (or variability), some of them being:
- Common motifs present in both sequences
- Likelihood of 2 sequences evolving from the same sequence
- Find the sequences from the Database similar to the sequence at hand
In this blog, I would like to step through some design and implementation details in sequence alignment using the Smith-Waterman algorithm (which is currently used in SSEARCH, OSEARCH and BLAST) to align a given biological sequence with a database of sequences and report the n-best fits.
Serial Approach:
The database consists of multiple sequences represented in FASTA format. All the sequences in FASTA format begins with a single-line description, followed by lines of sequence data. The description line is distinguished from the sequence data by a greater-than (">") symbol in the first column. The sequence ends if another line starting with a ">" appears; this indicates the start of another sequence. A simple example of one sequence in FASTA format is given below:
>gi|5524211|gb|AAD44166.1| cytochrome b [Elephas maximus maximus]
LCLYTHIGRNIYYGSYLYSETWNTGIMLLLITMATAFMGYVLPWGQMSFWGATVITNLFSAIPYIGTNLV
EWIWGGFSVDKATLNRFFAFHFILPFTMVALAGVHLTFLHETGSNNPLGLTSDSDKIPFHPYYTIKDFLG
LLILILLLLLLALLSPDMLGDPDNHMPADPLNTPLHIKPEWYFLFAYAILRSVPNKLGGVLALFLSIVIL
GLMPFLHTSKHRSMMLRPLSQALFWTLTMDLLTLTWIGSQPVEYPYTIIGQMASILYFSIILAFLPIAGX
IENY
With the dependency stated above, the requirement was for the database file to be parsed sequentially and to process the records by concatenating each line except the header, till we reach the next record's header or the end of the file. Each of the processed records need to be aligned with the given subject sequence. The high-level serial approach is given below:
Read File and Process Record
For every record
Align Database Query (record) with Subject (input)
Update output vector
Parallel Approach:
Agreed, there is a dependency in reading the file sequentially to obtain the query sequences, however, we can align the sequences in any order we want. This makes it an embarrassingly parallel problem.
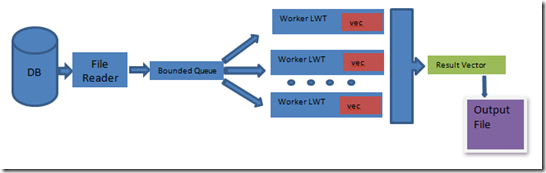
The approach is to implement a typical producer-consumer scenario, with 1 producer (file reader) and n consumers (n being the processor count). The producer reads the file and process records and enqueues it into a bounded concurrent queue. Once the producers are done processing the database file, they set their done flag. The consumers on the other end of the bounded concurrent queue dequeue each record and align them using the Smith-Waterman approach. The consumers then update their local vector (which holds a max m-user specified number of elements) with the results, until the producer's done flag is set and the queue is empty. All the local vectors and then combined into a result vector which is sorted and truncated and is then written to an output file.
Initially, a concurrent queue was used instead of a bounded concurrent queue, but considering the nature of the problem where the file reader overwhelmed the queue with records which resulted in the system running out of resources (out of memory), the decision was made to have an upper bound beyond which the producer will cooperatively block to enhance forward progress. The ratio of time for producer:consumers was approximately 1:25 and the file reader ended up polling for a static queue counter to decide if it had to process more records or yield (and let the consumers catch up).
When analyzing the implementation of the above approach using PPA, memory allocation stood out as the hotpath in the summary view.
This was a direct result of allocating and de-allocating the arrays in the smith-waterman class for every record, by every consumer. This was overcome by caching and reusing pre-allocated memory by each consumer context. The Smith-Waterman algorithm invoked by the consumers (with memory caching/reuse) is given below:
1: #pragma once
2:
3: #include "stdafx.h"
4: #include "ScoringMatrix.h"
5: #include "Alignment.h"
6: #include "Result.h"
7:
8: using namespace std;
9: using namespace Concurrency;
10:
11: #pragma warning(disable:4018)
12: #pragma warning(disable:4267)
13:
14: class Smith_Waterman
15: {
16: public:
17: /// <summary>
18: /// Ctor used when searching a Database
19: /// </summary>
20: ///
21: /// <param name="subject">The subject to align<param>
22: /// <param name="maxQueryLength">A preset max query length<param>
23: /// <param name="matrix">The scoring matrix to use<param>
24: Smith_Waterman(string subject, unsigned int maxQueryLength, ScoringMatrixBase* pMatrix)
25: :m_subjectLength(subject.length()),
26: m_maxQueryLength(maxQueryLength),
27: m_pMatrix(pMatrix)
28: {
29: m_pSubject = new char[m_subjectLength+1];
30: strcpy_s(m_pSubject, m_subjectLength+1, subject.c_str());
31:
32: m_pQuery = new char[m_maxQueryLength+1];
33:
34: m_ppDirectionMatrix = new unsigned char*[m_subjectLength];
35: for(int i=0; i < m_subjectLength; ++i)
36: {
37: m_ppDirectionMatrix[i] = new unsigned char[m_maxQueryLength];
38: memset(m_ppDirectionMatrix[i], 0, m_maxQueryLength * sizeof(unsigned char)); //Set to 0's
39: }
40:
41: m_pLocalAlignScore_E = new int[m_maxQueryLength];
42: memset(m_pLocalAlignScore_E, 0, m_maxQueryLength * sizeof(int));
43:
44: m_pMaxLocalAlignScore_H = new int[m_maxQueryLength];
45: memset(m_pMaxLocalAlignScore_H, 0, m_maxQueryLength * sizeof(int));
46: }
47:
48:
49: ~Smith_Waterman()
50: {
51: delete [] m_pSubject;
52: delete [] m_pQuery;
53: delete[] m_pLocalAlignScore_E;
54: delete[] m_pMaxLocalAlignScore_H;
55: for (int i = 0; i < m_subjectLength; ++i)
56: {
57: delete m_ppDirectionMatrix[i];
58: }
59: delete [] m_ppDirectionMatrix;
60: }
61:
62: /// <summary>
63: /// On analysis (PPA), 1 of the bottlenecks around parallelism was around
64: /// HeapLock (contention). The approach has been revised to minimize allocation/reallocation
65: /// by object reuse.
66: /// </summary>
67: ///
68: /// <param name="query">The query string from parsing the db<param>
69: void SetQuery(string query)
70: {
71: if (query.length() > m_maxQueryLength)
72: {
73: //Free memory and reallocate
74: m_maxQueryLength = query.length();
75: delete [] m_pQuery;
76: m_pQuery = new char[m_maxQueryLength+1];
77:
78: delete[] m_pLocalAlignScore_E;
79: m_pLocalAlignScore_E = new int[m_maxQueryLength];
80:
81: delete[] m_pMaxLocalAlignScore_H;
82: m_pMaxLocalAlignScore_H = new int[m_maxQueryLength];
83:
84: for (int i = 0; i < m_subjectLength; ++i)
85: {
86: delete m_ppDirectionMatrix[i];
87: m_ppDirectionMatrix[i] = new unsigned char[m_maxQueryLength];
88: }
89: }
90:
91: m_queryLength = query.length();
92: memset(m_pQuery, '\0', (m_queryLength+1) * sizeof(char));
93: strcpy_s(m_pQuery, m_queryLength+1, query.c_str());
94:
95: for (int i = 0; i < m_subjectLength; ++i)
96: {
97: memset(m_ppDirectionMatrix[i], 0, m_queryLength * sizeof(unsigned char)); //Set to 0's
98: }
99: memset(m_pLocalAlignScore_E, 0, m_maxQueryLength * sizeof(int));
100: memset(m_pMaxLocalAlignScore_H, 0, m_maxQueryLength * sizeof(int));
101:
102: assert(m_maxQueryLength >= m_queryLength);
103: }
104:
105:
106: /// <summary>
107: /// Used when aligning Query
108: /// This calls Popoluate followed by Traceback
109: /// </summary>
110: ///
111: /// <param name="gap_open">const cost to open gap<param>
112: /// <param name="gap_extn">const cost to extend gap<param>
113: /// <param name="fRecursiveTraceback">Trace all paths/mutations<param>
114: Result Align(int gap_open, int gap_extn, const bool fRecursiveTraceback)
115: {
116: return TraceBack(Populate(gap_open, gap_extn), fRecursiveTraceback);
117: }
118:
119: /// <summary>
120: /// Used when aligning Query
121: /// This calls Popoluate followed by Traceback
122: /// </summary>
123: ///
124: /// <param name="gap_open">const cost to open gap<param>
125: /// <param name="gap_extn">const cost to extend gap<param>
126: Element Populate(int gap_open, int gap_extn)
127: {
128: Element elem;
129: int row, col, localAlignScore_F, diagonalMaxLocalAlignScore_H, localAlignScore_H;
130: for (row = 1; row < m_subjectLength; ++row)
131: {
132: localAlignScore_F = 0;
133: diagonalMaxLocalAlignScore_H = m_pMaxLocalAlignScore_H[0];
134: for (col = 1; col < m_queryLength; ++col)
135: {
136: //Calc local alignment (match/mismatch) score: H(i-1, j-1) + SimilarityScore(A[i], B[j])
137: localAlignScore_H = diagonalMaxLocalAlignScore_H + m_pMatrix->GetScore(m_pSubject[row-1], m_pQuery[col-1]);
138:
139: //Calc deletion score
140: m_pLocalAlignScore_E[col] = max((m_pLocalAlignScore_E[col] - gap_extn), (m_pMaxLocalAlignScore_H[col] - gap_open));
141:
142: //Calc insertion score
143: localAlignScore_F = max((localAlignScore_F - gap_extn), (m_pMaxLocalAlignScore_H[col - 1] - gap_open));
144:
145: //Store away the diag score
146: diagonalMaxLocalAlignScore_H = m_pMaxLocalAlignScore_H[col];
147:
148: //Calculate matrix value h(i,j) = max{(h(i-1, j-1) + Z[A[i], B[j]]), e(i,j), f(i,j), 0}
149: m_pMaxLocalAlignScore_H[col] = MAX4(localAlignScore_H, m_pLocalAlignScore_E[col], localAlignScore_F, 0);
150:
151: // Determine the traceback direction
152: (m_pMaxLocalAlignScore_H[col] == localAlignScore_H) ? m_ppDirectionMatrix[row][col] += DIAG : __noop;
153: (m_pMaxLocalAlignScore_H[col] == m_pLocalAlignScore_E[col]) ? m_ppDirectionMatrix[row][col] += TOP: __noop;
154: (m_pMaxLocalAlignScore_H[col] == localAlignScore_F) ? m_ppDirectionMatrix[row][col] += LEFT : __noop;
155:
156: // Set the traceback start at the current elem i, j and score
157: (m_pMaxLocalAlignScore_H[col] > elem.GetScore()) ? elem.Set(row, col, m_pMaxLocalAlignScore_H[col]) : __noop;
158: }
159: }
160: return elem;
161: }
162:
163: /// <summary>
164: /// Used when Walkback the matrix from the winning node
165: /// </summary>
166: ///
167: /// <param name="elem">Winning element<param>
168: /// <param name="fRecursiveTraceback">Trace all paths/mutations<param>
169: Result TraceBack(Element elem, const bool fRecursiveTraceback)
170: {
171: int row = elem.GetRow();
172: int col = elem.GetColumn();
173: Result result(m_pSubject, m_pQuery, elem.GetScore());
174: NodeWalk(result, row, col, fRecursiveTraceback);
175: return result;
176: }
177:
178: private:
179:
180: /// <summary>
181: /// Walk back from the winning node based on
182: /// direction set during populating matrix
183: /// </summary>
184: ///
185: /// <param name="result">computed Result value<param>
186: /// <param name="row">The row from where nodewalk needs to be performed<param>
187: /// <param name="col">The col from where nodewalk needs to be performed<param>
188: /// <param name="fRecursiveTraceback">Trace all paths/mutations<param>
189: /// <param name="fFirstPass">Oneshot effective only when fRecursiveTraceback is true<param>
190: /// <param name="alignedSubject">reversed subject string, value determined by the direction<param>
191: /// <param name="markup">relation between alignedSubject and alignedQuery chars<param>
192: /// <param name="alignedQuery">reversed query string, value determined by the direction<param>
193: void NodeWalk(Result& result, int row, int col, const bool fRecursiveTraceback, const bool fFirstPass=true, string alignedSubject="", string markup="", string alignedQuery="")
194: {
195: unsigned char trace;
196: bool fTerminate=false;
197: if (fFirstPass)
198: {
199: alignedSubject.push_back(m_pSubject[row]);
200: alignedQuery.push_back(m_pQuery[col]);
201:
202: if (m_pSubject[row] == m_pQuery[col])
203: {
204: markup.push_back(IDENTITY);
205: result.m_identity++;
206: result.m_similarity++;
207: }
208: else if (m_pMatrix->GetScore(m_pSubject[row], m_pQuery[col]) > 0)
209: {
210: markup.push_back(SIMILARITY);
211: result.m_similarity++;
212: }
213: else
214: {
215: markup.push_back(MISMATCH);
216: }
217: }
218:
219: while(!fTerminate)
220: {
221: if((m_ppDirectionMatrix[row][col] & DIAG) != 0)
222: {
223: trace = DIAG;
224: alignedSubject.push_back(m_pSubject[row-1]);
225: alignedQuery.push_back(m_pQuery[col-1]);
226: if (m_pSubject[row-1] == m_pQuery[col-1])
227: {
228: markup.push_back(IDENTITY);
229: result.m_identity++;
230: result.m_similarity++;
231: }
232: else if (m_pMatrix->GetScore(m_pSubject[row-1], m_pQuery[col-1]) > 0)
233: {
234: markup.push_back(SIMILARITY);
235: result.m_similarity++;
236: }
237: else
238: {
239: markup.push_back(MISMATCH);
240: }
241:
242: if (m_ppDirectionMatrix[row][col] != trace)
243: {
244: //More than 1 path exists
245: m_ppDirectionMatrix[row][col] -= DIAG;
246: (fRecursiveTraceback) ? NodeWalk(result, row, col, true, false, alignedSubject, markup, alignedQuery) : __noop;
247: }
248: --row;
249: --col;
250: continue;
251: }
252:
253: if((m_ppDirectionMatrix[row][col] & TOP) != 0)
254: {
255: trace = TOP;
256: alignedSubject.push_back(m_pSubject[row-1]);
257: alignedQuery.push_back('-');
258: markup.push_back(GAP);
259: result.m_gaps++;
260: if (m_ppDirectionMatrix[row][col] != trace)
261: {
262: //More than 1 path exists
263: m_ppDirectionMatrix[row][col] -= TOP;
264: (fRecursiveTraceback) ? NodeWalk(result, row, col, true, false, alignedSubject, markup, alignedQuery) : __noop;
265: }
266: --row;
267: continue;
268: }
269:
270: if((m_ppDirectionMatrix[row][col] & LEFT) != 0)
271: {
272: trace = LEFT;
273: alignedSubject.push_back('-');
274: alignedQuery.push_back(m_pQuery[col-1]);
275: markup.push_back(GAP);
276: result.m_gaps++;
277: if (m_ppDirectionMatrix[row][col] != trace)
278: {
279: //More than 1 path exists
280: m_ppDirectionMatrix[row][col] -= LEFT;
281: (fRecursiveTraceback) ? NodeWalk(result, row, col, true, false, alignedSubject, markup, alignedQuery) : __noop;
282: }
283: --col;
284: continue;
285: }
286:
287: if(m_ppDirectionMatrix[row][col] == 0)
288: {
289: //Terminating condition
290: result.m_alignments.push_back(Alignment(alignedSubject, markup, alignedQuery));
291: fTerminate=true;
292: }
293: else
294: {
295: throw;
296: }
297: }
298: }
299:
300: private:
301: unsigned char** m_ppDirectionMatrix;
302: char* m_pSubject;
303: char* m_pQuery;
304: const unsigned int m_subjectLength;
305: unsigned int m_queryLength;
306: int* m_pLocalAlignScore_E;
307: int* m_pMaxLocalAlignScore_H;
308: unsigned int m_maxQueryLength;
309: ScoringMatrixBase* m_pMatrix;
310: };
311:
312: #pragma warning(default:4018)
313:
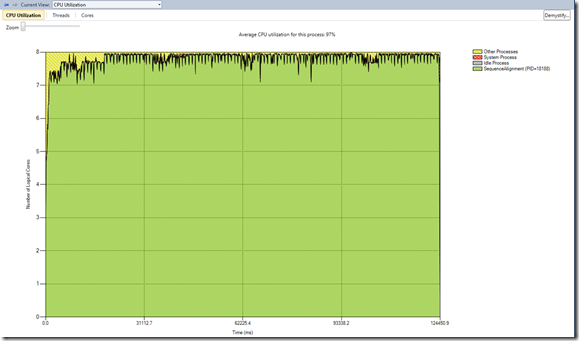
With that implemented, the next step was to check the cpu utilization under-utilized processors. From the view below, we notice that the average cpu utilization was 97% and that there is some minor idle process (3%)
- some idle process at the beginning of the parallelization - this is accounted to the initial part of the app before the consumers had work
- some at the end after the joining - this is accounted to the serialization of copy of the local array to the result array, sorting and truncating the result array and writing it to file.
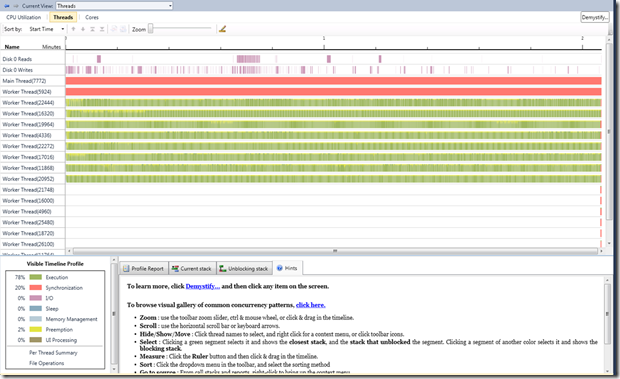
The thread view in PPA gives a more definitive picture on what was going on. Please note that this was recorded on an 8-core machine. From the view, you would observe that:
- there are 8 threads that are either executing or are preempted
- the main thread is blocked until the consumers complete
Additional notes:
- Thread 5924 happens to be a background scheduler thread, which basically gathers task statistics
- All other threads are introduced by the sampling routine
This seems like a healthy picture. Further analysis was done to ensure that there were no unnecessary/buggy spinning in user code. (This can also be inferred from the functions window)
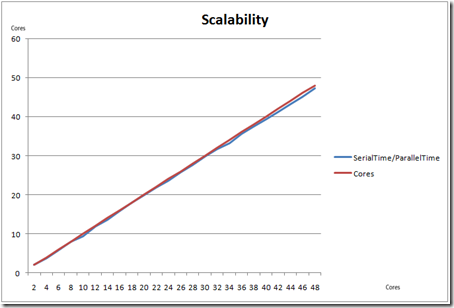
Results and Performance:
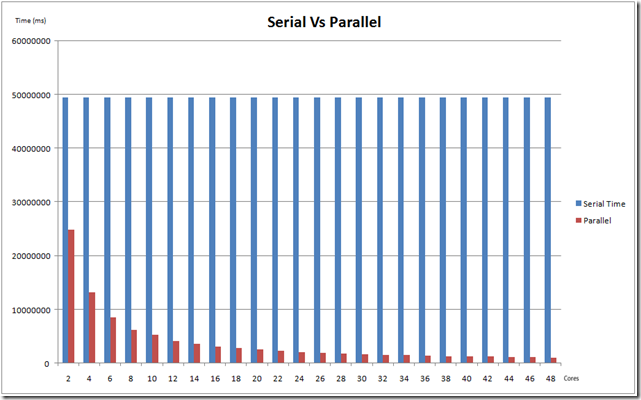
After analysis using PPA, this was application was run on a 48-way box. Given below are a set of timings comparing the parallel runs with the serial runs, using 2-48 cores (to address scalability).
| Cores | Parallel Time (in ms) | Serial Time (in ms) | Speedup |
| 2 | 24754934 | 49408452 | 1.995903201 |
| 4 | 13180057 | 49408452 | 3.748728249 |
| 6 | 8544194 | 49408452 | 5.782693136 |
| 8 | 6170857 | 49408452 | 8.006740717 |
| 10 | 5256198 | 49408452 | 9.4000363 |
| 12 | 4139975 | 49408452 | 11.93448076 |
| 14 | 3635331 | 49408452 | 13.59118386 |
| 16 | 3118978 | 49408452 | 15.84123133 |
| 18 | 2760319 | 49408452 | 17.89954422 |
| 20 | 2494642 | 49408452 | 19.80582865 |
| 22 | 2254536 | 49408452 | 21.9151311 |
| 24 | 2095044 | 49408452 | 23.58349133 |
| 26 | 1916544 | 49408452 | 25.7799727 |
| 28 | 1786685 | 49408452 | 27.65370057 |
| 30 | 1658796 | 49408452 | 29.78573134 |
| 32 | 1563398 | 49408452 | 31.60324626 |
| 34 | 1487879 | 49408452 | 33.20730516 |
| 36 | 1388026 | 49408452 | 35.59620065 |
| 38 | 1318315 | 49408452 | 37.47848731 |
| 40 | 1255547 | 49408452 | 39.35213258 |
| 42 | 1201544 | 49408452 | 41.12080124 |
| 44 | 1142711 | 49408452 | 43.23792455 |
| 46 | 1098964 | 49408452 | 44.95911786 |
| 48 | 1044838 | 49408452 | 47.2881461 |
The chart below compares the serial and parallel times for cores from 2-48
The key component here is scalability. It was observed that the application achieved close to linear scalability, when measured for 2-48 cores.
Summary:
To summarize, a scalable and simple fast-producer, slow-consumer type of problem was effectively parallelized using asynchronous scheduled tasks, cooperative synchronization primitives to enhance forward progress and a bounded concurrent queue (to block the producer if upper bound is reached). Visual Studio 2010 tools such as PPA was used for analyzing performance and the parallel stacks and parallel tasks windows (and the enhanced visualizers) were used for debugging the application.
Concrt Patterns used
- Bounded Concurrent Queue (concurrent queue + semaphore) - for asynchronous file processing
- Event - to signal completion of I/O reads
- Scheduler Primitives (Current Scheduler Create/ScheduleTask/Detach)