University of Oxford's Interdisciplinary Bioscience Doctoral Training Programme Data Hack
Guest blog by Ellen Pasternack, DPhil candidate at the University of Oxford
About me
Hi! I'm Ellen Pasternack, a DPhil candidate in the University of Oxford's Interdisciplinary Bioscience Doctoral Training Programme.
LinkedIn: https://www.linkedin.com/in/ellen-pasternack-6b1191125/
I’m studying sexual selection in red jungle fowl, and I also blog about biology at www.endlessformsmostwonderful.wordpress.com. As part of my training this autumn, I undertook a three week course in bioinformatics, led by Dr Phil Fowler from the Nuffield Department of Medicine.
Introduction
This year for the first time, Phil was trialling using Microsoft Azure as a tool for course. With Azure, we could create Bio-Informatics virtual machines from saved images set up by instructors with relevant programmes already installed, saving a lot of time that would otherwise be spent installing programmes on each individual computer, only then to be used once or twice. Azure also allowed us remote access to much more powerful machines than we were using in the department, which vastly sped up our work, given the massive datasets used in bioinformatics.
Biology Data Hacking
The final week of the course was structured as a hackathon: in groups, we'd put into practice techniques that we'd learned to solve novel biological problems of our choosing. My group set out to investigate the genetics of bird migration, following team leader Joe's passion for ornithology.
Our hackathon team, from left to right: Brandon Ford, myself, and Joe Wynn.
Ornithology Hack Project
Bird migration is a really fascinating phenomenon, which remains full of mysteries to this day. The fact that birds migrate was only settled in 1928— over a decade after Einstein's theory of general relativity was published. We now know that many species travel huge distances in astonishing feats of endurance- for instance, arctic terns fly up to 90,000km from pole to pole and back again every single year. We know that the routes of migration can be innate; birds often don't need to be taught where to go, but may have an instinctive drive to fly in a certain direction. The magnetic field of the Earth seems to be involved for some species, though not all. And these incredible cross-continental journeys haven't evolved just once, but crop up again and again in the evolutionary tree of life. Why? We don't know.
Our first thought was to use orthofinder (Emms and Kelly 2015), a piece of software we'd worked with in a prior practical, to see whether there were any homologous groups of proteins more commonly found in migratory birds compared to non-migratory species. On the NCBI website, we were able to find 62 complete avian proteomes to download and run through orthofinder. We went through and categorised each species according to the extent of its migration: from no migration to 'full migration', where the species has two distinct geographic ranges for summer and winter, plus a couple of in-between statuses, such as 'migration in some parts of range'. Our hope was that we'd be able to identify a gene that was more commonly found in migrants, that— fingers crossed— coded for something relevant to migration, such as fat metabolism.
At first, we waited excitedly for orthofinder to spit out its results. But when the next day came with very little indication of progress, we realised we had a problem. When we used orthofinder before, we'd been looking at bacteria, which have famously short genomes. Not only were the genomes of our bird species much longer than a typical bacterium, but there were more of them. Since the time taken to compare each individual to each other individual would scale factorially, the programme wouldn't finish running for anywhere from months to longer than the universe has existed. Time for a change of plan.
Joe then thought back to a paper published earlier in 2017, in which researchers identified four genes that were upregulated in blackbirds before and during migration (Franchini et al. 2017). Our plan B was to look at these particular genes, and see whether migration was associated with a difference in their sequence as well as expression rate.
The four genes we looked at were:
· DNA topoisomerase 2— this fixes errors in DNA replication, as well as regulating chromatin condensation. These processes are very important in cellular division.
· TGF beta receptor type 1— this is a cytokine involved in stimulating cell growth in a wide variety of tissue types.
· ST6GALNAC2, which we affectionately named 'stgnac'— this gene codes for an enzyme with a very long unpronounceable name, involved in breaking down energy storage molecules so they can be used.
· A motilin receptor— motilin is a peptide that stimulates smooth muscle contraction in the small intestine.
All of these genes are quite general-purpose, important genes that are found in many species. However, it’s possible that they may be even more important for migratory birds. Birds often undergo considerable physiological changes prior to migration, replacing all their feathers and building up flight muscles and fat reserves to sustain them over the strenuous journey ahead. Since migratory birds must go through this period of rapid growth, as well as intense endurance, these genes relating to general bodily activity may have been selected differently in migratory species than non-migratory ones.
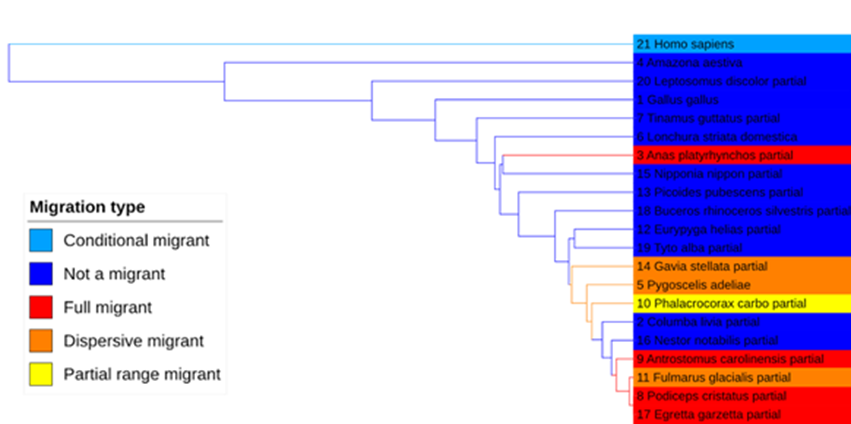
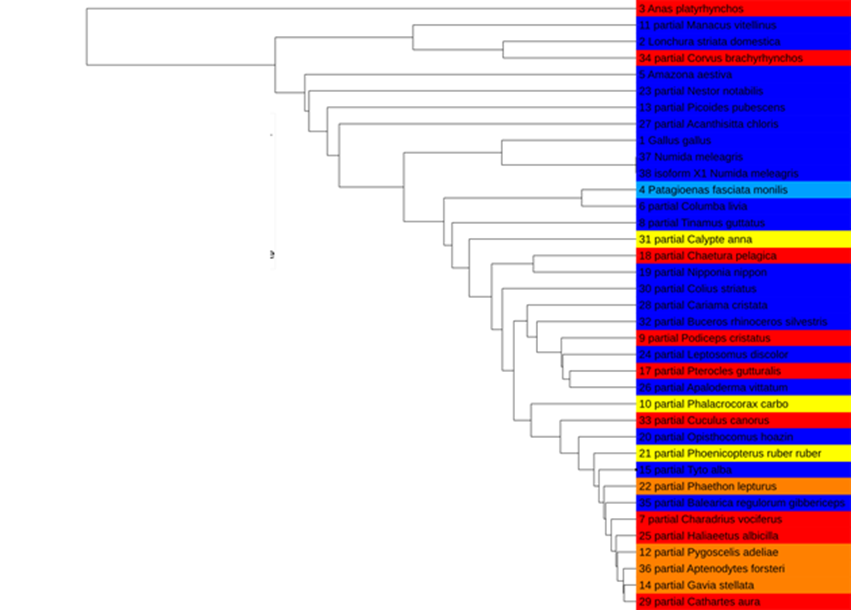
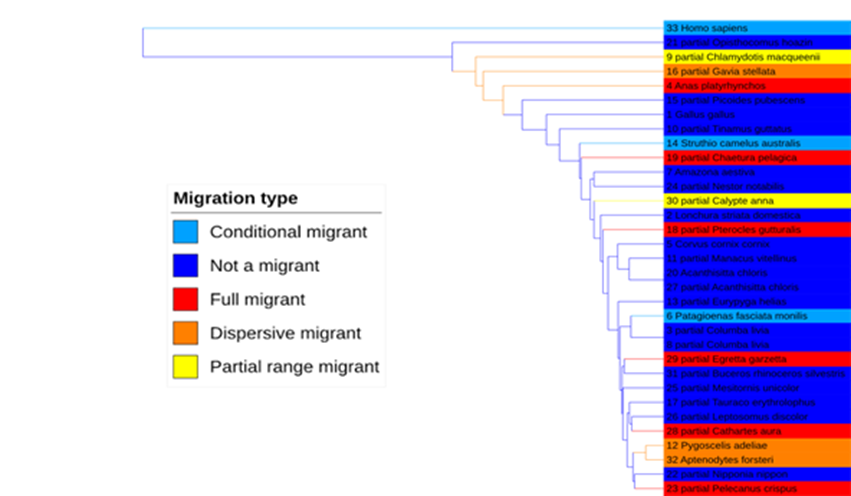
The next step was to download the protein sequences for these genes for every bird species available from NCBI, as well as for humans as an outgroup. There weren’t enough sequences available for the motilin receptor to provide any meaningful insight, but for the remaining three genes, we used MAFFT to align the sequences and produce phylogenetic trees based on each. These trees are visualised below using the Interactive Tree Of Life or iTOL, a web-based tree builder.
DNA topoisomerase 2b:
TGF-beta receptor type 1:
ST6GALNAC2:
As you can see, each gene produces a slightly different phylogeny, and gratifyingly, humans are correctly placed as the outgroup in each analysis. The topoisomerase tree puts migrants as loosely clustered together, which might indicate some degree of convergent evolution of this gene in migrant species. We don’t see this in the trees based on the other two genes, however.
We then wanted to quantify how conserved the protein sequences of these genes might be in migratory species, compared to non-migrants. To do this, we used a Needleman-Wunsch algorithm to generate similarity scores for each possible two-way comparison between migratory species’ sequences. We then repeated this for non-migrants, and performed t-tests comparing the level of difference observed between migratory species and between non-migratory species for each gene. (Humans were left out at this stage, because as an outgroup they would skew the results towards more difference in non-migrants).
This methodology is slightly unorthodox, because we are comparing our results to a null model where the pairwise difference scores between sequences is randomly distributed, but really we should be using a null model that takes into account a baseline rate of sequence divergence, so that species more closely related to each other are expected to have more similar protein sequences. Additionally, our method treated each individual comparison as an independent data point, which may have led to pseudoreplication and misrepresentation of the data if, for instance, there was one particularly outlying sequence which would then generate high difference scores with every other sequence it was compared to. There definitely are better methods that we could have used, but we didn’t manage to look into them- unfortunately, we were running very short on time at this point due to our complete change of direction on the second day. Our statistical analysis is therefore very rough around the edges, but we hoped we could get away with it since migration is fairly evenly distributed in bird phylogeny, so there wasn’t much in the way of underlying structure that was being ignored.
With that being said, we did find a very significant test result for all three of the genes we looked at, with migrant species showing much more similarity between sequences than non-migrants did. Our very tentative conclusion was therefore that these genes which were upregulated in migrants were also more closely conserved in migratory species. However, this has yet to stand up to more stringent statistical scrutiny, so could still turn out to be totally wrong! Another thing we’d be interested in looking into further is whether there are any amino acid substitutions, insertions or deletions in particular in these sequences that are associated with migratory species, rather than this very rough measure of general similarity.
Presenting our findings
On the last day everybody presented their results. It was very enjoyable hearing about what the other teams had got up to; including looking for vaccine targets in a virus genome, investigating the molecules involved in photosynthesis, and simulating a molecule passing through a nanopore. My teammates and I were delighted to be voted third place in the hackathon.
Conclusion
None of us in my team had a background in computing— in fact, we'd only been introduced to Linux a few weeks earlier. All of us were very new to this. Despite this, we found Microsoft Azure and the Bioinformatics Virtual Machine a straightforward way of working together, sharing files and collaborating on tasks across multiple monitors.
Next Steps
We were far less constrained in the scope of our project than we would have been if we were restricted to the much smaller processing power of the departmental desktops. I would highly recommend Azure and custom Virtual Machine as a way of working on collaborative projects, especially with the very large datasets that are often involved in bioinformatics.