Novel HoloLens Application - Genetic Data Visualisation in Mixed Reality
Guest post by Rachel Slater, M.Sc Student at UCL, where I am taking courses such as Algorithmic's and Artificial Intelligence & Neural Computing. I am the Community Lead at London's She++ chapter, an undergraduate teaching assistant at UCL, and a Faculty StAR (student academic representative). Prior to UCL, I completed a Medicinal Chemistry degree at Trinity College Dublin, followed by a Product Management position at a Dublin-based technology company. I received a full scholarship as a 'promising Woman in Tech' in 2015 and have spent over 6 months in the San Francisco Bay Area, attending the Coding House Institute's programme.
I love to build things, and have a particular interest in applying technology to Medicine and Health.
My MSc Project
Having spent this past summer immersed in HoloLens development, I’m now in the exciting phase of communicating the experience and results, and presenting the final application.
As a M.Sc Computer Science student at UCL, I got the opportunity to work on an experimental project which aligned perfectly with my interest in applying advanced technology to medicine and healthcare.
The past 12 weeks have culminated in a novel application for the HoloLens device. The application, and supporting thesis, is titled Genetic Data Visualisation in Mixed Reality. Being my first experience with MR/AR development, the project involved learning C# programming, Unity development, and working with REST APIs. It’s the first HoloLens application of its kind - a search engine for genetic data in mixed reality.
The primary objective of this project was to create a genome browser - a crucial tool used in the daily work of thousands of scientists and physicians to search, retrieve and analyse genomic data, in an analogous way to navigating the internet - in mixed reality. Genome browsers are used to visualise both genomic data and genomic features.
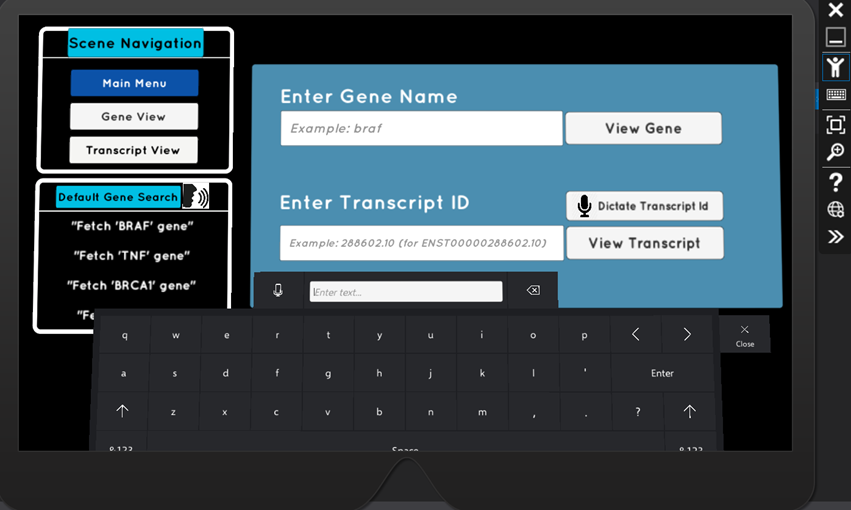
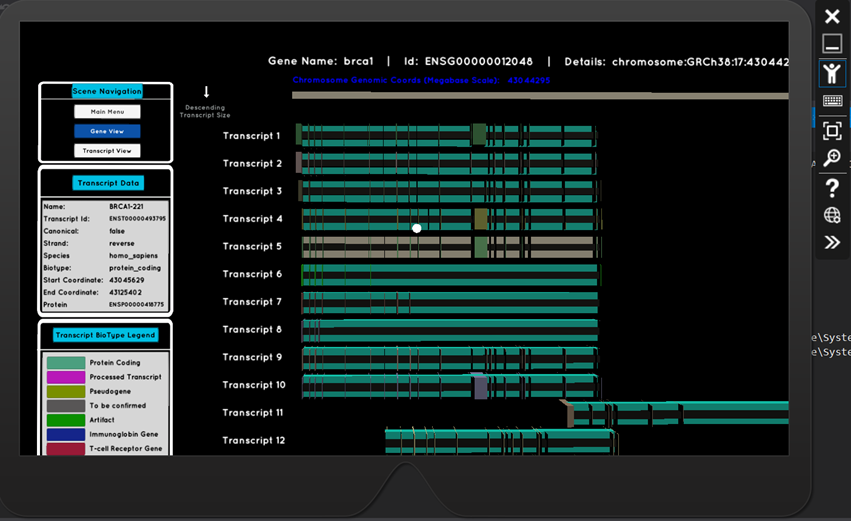
The resulting HoloLens application delivers on both of these fronts. Ensembl, a leading organisation in genomics data (funded by the Wellcome Trust Sanger Institute and the European Bioinformatics Institute), have a REST API, which is now a key part of this application. Integration with this API, means that any gene name or transcript id search input can be handled. The retrieved data is used to generate 3D visualisations which are scientifically precise - rendered transcripts are scaled and positioned based on their genomic coordinates along the chromosomes. Exon and intron positions are calculated algorithmically, and are rendered in their respective locations along the transcript.
Full HoloLens capabilities have been developed, making the experience interactive. Users can search for genetic data using voice commands, or via a gesture-controlled keyboard hologram.
Their gaze, focussed on a particular transcript, triggers the appearance of a data panel displaying its characteristic information.
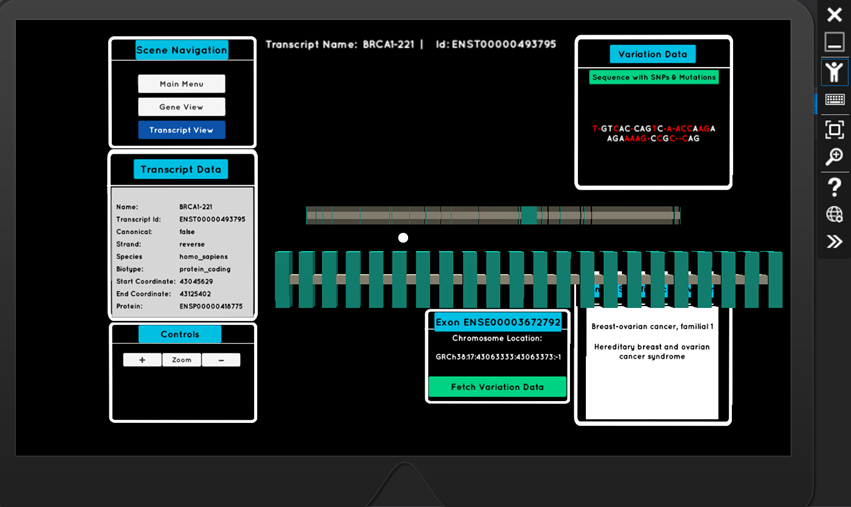
The air-tap gesture, when targeting a transcript, takes the user to a new scene where they can delve deeper into the exons (the important coding regions of DNA). Again, through interactive holograms, they can explore an exon’s sequence of nucleotides, it’s common mutations (SNPs - single nucleotide polymorphisms), and associated phenotypes which are of clinical significance (for example, heart disease)
What began as an experimental project, produced a robust and practical application which has been validated by rigorous rounds of testing.
On September 8th, I got the chance to present the project to the HoloLens team in their studio at the Microsoft London offices.
Getting face-to-face feedback from the team was fantastic, as, naturally, being experts in the technology, they spotted things that other eyes couldn’t. I began by explaining the concept and my approach, followed by a HoloLens demo and feedback session. The team provided invaluable tips, particularly on a couple of remaining bugs and nuisances in the app that I hadn’t solved.
Overall, the project has been challenging and exciting, in equal measure. To have built an application that provides information down to the level of DNA sequence, mutation alleles, and phenotype traits, all in mixed reality, is a result I’m delighted with, and I plan to submit the app to the Windows Store in the coming weeks.
A video of the MR experience, filmed at London’s Wellcome Trust, can be seen here.
I’d love to hear your comments and feedback you can learn more about the PEACH projects at https://medium.com/ucl-peach/first-steps-peach-reality-genomics-and-proteomics-caceb1af685a
Resources
Homepage:
Hardware:
https://www.microsoft.com/microsoft-hololens/en-gb/development-edition#de-accordion-tech-spec-panel
Commercial Suite (and comparison to DevEdition):
https://www.microsoft.com/microsoft-hololens/en-gb/commercial-suite
https://developer.microsoft.com/en-gb/windows/holographic/release_notes
https://developer.microsoft.com/en-gb/windows/holographic/commercial_features
Purchase Devices